With a New ‘Map’ of Cells, Georgetown Cancer Researchers Chart a Course to More Effective Treatment
Among the many challenges of effective cancer treatment is the need to monitor the cancer itself. Keeping track of cancer cells as they grow, and hopefully as they die off, may be inexact, too infrequent, and expensive for patients.
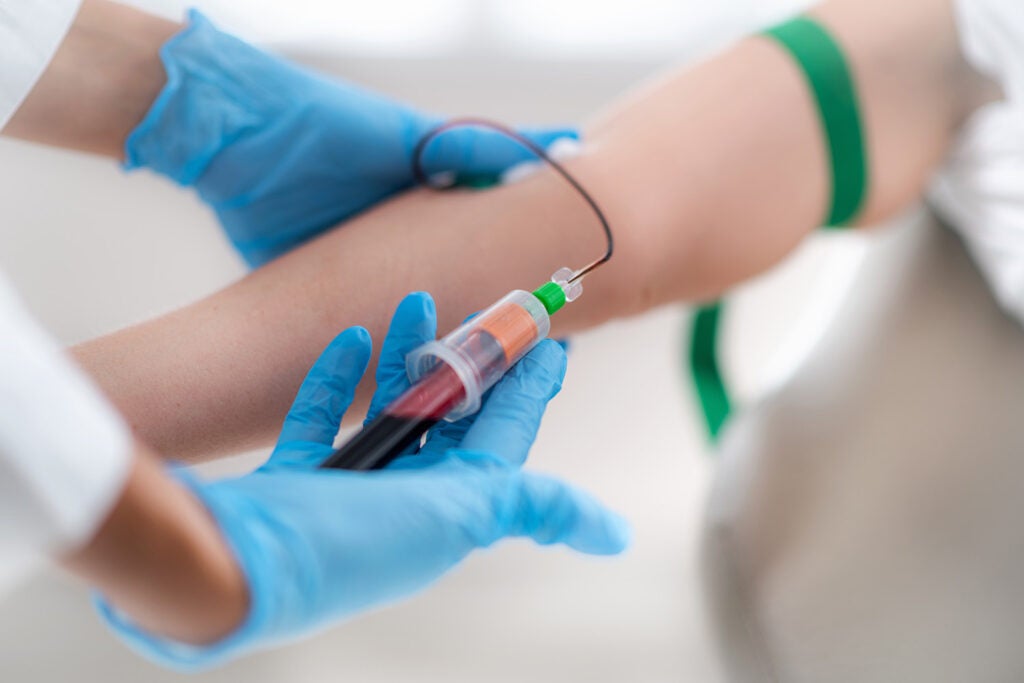
A group of Georgetown University Medical Center (GUMC) researchers wants to change that. Faculty and students from Biomedical Graduate Education programs, the School of Medicine and Lombardi Comprehensive Cancer Center are developing a new technique that could revolutionize the way cancers are tracked and treated. Instead of invasive surgical biopsies or time-consuming scans, clinicians could use simple blood tests to determine where the cancer is and whether it is responding to treatment.
“From a blood sample, we think you can detect whether or not a drug works earlier – much, much earlier,” said Anton Wellstein, an oncology and pharmacology professor who is leading the research. “We also see the side effects the drug has at the same time.”
Bits in the Bloodstream
The researchers are working with the relatively young technology of liquid biopsy, which involves examining normal and cancer cell DNA in the bloodstream. When cells die, some of their DNA ends up floating through the circulatory system. Advances in the past decade allow labs to test this loose DNA for evidence of cancer mutations, and in recent years to trace normal cell DNA to its tissue of origin.
The presence of mutated DNA in the bloodstream can be an early warning sign that a tumor is growing and sloughing off cells. During treatment, mutant DNA levels in the blood should spike, then disappear as the tumor dissolves.

Anton Wellstein
“It’s a more objective measure” compared to an X-ray or MRI image of a tumor, Wellstein said. “The beauty of the analysis is really that you can get insight into what happens” during treatment.
A blood test doesn’t necessarily provide a simple picture. DNA in blood can come from anywhere in the body, and clinicians need to know which organs are diseased in order to treat them.
At Georgetown, researchers are compiling a “map” of cell types that can be used to tell which organs that DNA is coming from – providing a clearer picture that can guide rapid, personalized treatment.
“There are hundreds of cell types in the body, as far as we know – it depends on how deep you want to dig,” Wellstein said. “The map that we currently have is about 40 … major cell types in the body. … Forty is quite a few when you try to go through it.”
Descended from DREAMseq
The mapping project is on a long journey from the clinic to the lab and back again. Its story begins with the nationwide DREAMseq clinical trial led by Michael Atkins, professor and deputy director of the Lombardi Comprehensive Cancer Center. “DREAMseq” stands for “Doublet, Randomized Evaluation in Advanced Melanoma Sequencing.”

Michael Atkins
In 2015, Atkins and collaborators started to evaluate the effectiveness of different treatments for metastatic BRAF-mutant melanoma. Some patients were started on targeted therapy, which uses drugs to attack vulnerable proteins essential to the cancer cells. Others began with immunotherapy treatment, which restores the ability of the patient’s immune system to clean up the cancer. Patients who responded poorly to one form of treatment were switched to the other.
The results were so striking that the trial was closed to accrual in fall 2021 by its data safety monitoring committee (DSMC).
“[The DSMC] and the National Cancer Institute looked at the data in an unblinded fashion and saw that there was a clinically meaningful 20 percent difference in two-year overall survival, favoring the group of people who got immunotherapy first,” Atkins explained. “Seventy-two percent of people who received immunotherapy first were alive, versus 52 percent of those who received targeted therapy.”
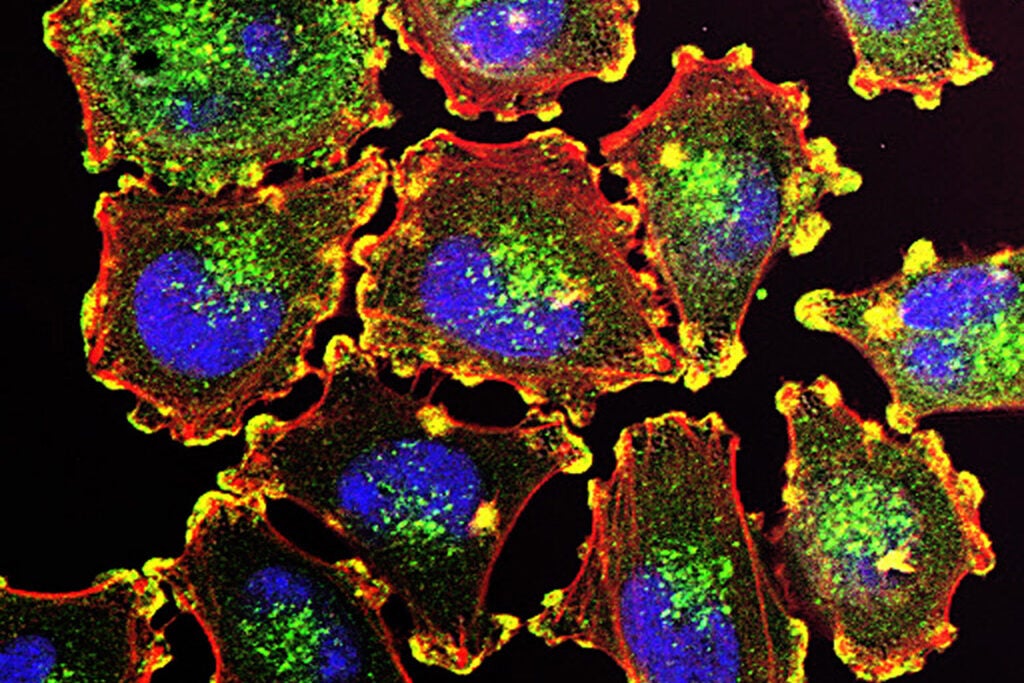
Metastatic melanoma cells. Courtesy of Julio C. Valencia, NCI Center for Cancer Research
The DREAMseq trial had made a leap forward in treating melanoma. The publication of its results was named 2023’s Paper of the Year in the Journal of Clinical Oncology.
“It’s already made a difference,” Atkins said. “It’s changed the treatment of these patients worldwide.”
Researchers are now using blood and tumor samples collected during the trial to learn more about how the treatments work. “We call that ‘reverse translation,’ where we go from observations in the clinic to the laboratory to help explain them and clarify them,” Atkins said. “And we may learn from those observations new things to actually test in the clinic, because new questions may be raised.”
Questions and challenges remained. A subset of patients had better outcomes when starting with targeted therapy, defying the trend. And in any case, clinicians needed to assess patients’ response to treatment as quickly as possible in order to set them on the best course.
Liquid biopsy was a possible solution. More Georgetown scientists stepped up to find out how well it could work.
“True Team Science”
Using blood and tissue samples collected during the DREAMseq trial, researchers in Wellstein’s lab started to build their map of cell types. The key to the map is DNA methylation patterns, epigenetic characteristics which guide cells in different organs to behave differently. The researchers found that when melanocyte skin cells mutate into melanoma, they tend to retain their signature methylation pattern.
Sidharth Jain sits at a workstation in the lab.
“We also found along the way that cancer changes and messes up so many things,” said Sidharth Jain (G’25 M’27), an M.D./Ph.D. in Tumor Biology student working in Wellstein’s lab. “Methylation is one of the things that gets messed up pretty frequently in cancer. But, we were lucky that the majority of the DNA methylation [that] defines those melanocytes … is still reflected in most melanoma.”
Jain said the researchers hope that methylation patterns could be used to identify other types of cancer cells as well: “We think that that’s mainly the case across most cancers. Cancers will retain their memory of being a normal cell.”
The complexity of the task – hundreds of samples, terabytes of genetic data – has brought together students and faculty from many parts of the medical center.
A succession of doctoral and master’s students from the Tumor Biology Program have played central roles in the project. As one of the latest predoctoral participants, Jain works at the bench and the computer to process data from patient blood samples. He extracts DNA from serum, enriches regions of interest to send for sequencing, then analyzes the data to determine which organs the DNA came from. It can take two 12-hour days to get through a single sample.

Patrick McDeed
Patrick McDeed (G’25), a Ph.D. in Biostatistics student, worked with Jain to develop the computational method the researchers use to analyze the samples. He explained: “Our job computationally is to learn some features that distinguish one fragment from another, to say, does this come from this cell type or tissue type versus another? And to trace those fragments back to the cell or tissue that they originated from – and then track those changes over time.”
The integration of wet lab, biostatistics and informatics work, as well as clinical insights, has forged a strong team and given students experience across disciplines.
“All the analyses that we’ve done in [Wellstein’s lab] have been really collaborative in that research design process,” McDeed said. “Which I think is really helpful, because a biologist views experimental design a certain way and data collection a certain way … and then I as a biostatistician have a different way of viewing that design and data collection and methods.
“It’s helpful to have all those conversations early on and have kind of that shared language and communication and true team science effort. And I think that’s led to some great collaboration and, hopefully, really interesting and impactful work further down the line – not only on this project, but other application areas that the lab is working on as well.”
The research has also been buoyed by colleagues around the world, including Atkins’ collaborators on the clinical trial as well as a group from Hebrew University in Jerusalem that contributed informatics experience.
“It’s a lot of group effort,” Wellstein said.
Shaping the Future
The researchers hope that their findings will be applied in clinical trials and lead to improvements in cancer care.

Sidharth Jain holds a bag of sample tubes in the lab.
“We want to publish it and apply it,” Wellstein said. “What we’re trying to do is also then make that available broadly. … Probably we have to commercialize it, have to find a company that will do the assays, because we won’t be able to do it in the lab.”
For the students of the lab, their role in the research is already influencing their professional futures. Megan McNamara (G’24, M’26), a Tumor Biology M.D./Ph.D. student who set up the mapping project during her Ph.D., is now completing her medical degree at the School of Medicine. Jain is on track to defend his dissertation in January 2025 before returning to the medical school as well. McDeed is getting ready to graduate and will be a postdoc at Fred Hutchinson Cancer Center in Seattle, a destination he said “very much came from all of this work.”
“The liquid biopsy space is not a huge field,” McDeed said. “It’s very promising emerging work. … There’s a few labs and research centers that are developing methods around this. [This project] opened up those doors to me.”
Read More

News, Student Profiles
Tumor Biology Student Sidharth Jain Combines Discovery and Care on M.D./Ph.D. Path
Sidharth Jain hopes that his experience at the lab bench and the bedside will help him to be an insightful researcher and compassionate clinician. As part of a melanoma research group, he is already working to make a difference in the diagnosis and treatment of cancer.
January 13, 2025

News
MedStar Georgetown University Hospital is the first hospital in the Washington metropolitan area to offer Tumor-Infiltrating Lymphocyte therapy, or TIL therapy, to treat metastatic melanoma.
January 8, 2025
News
The findings, coupled with results from another recent clinical trial, mark significant advances in the treatment of melanoma.
September 28, 2022

